Environmental DNA: the primary technology deployed in the framework of Vigilife
What is environmental DNA?

DNA is a molecule that is common to all living beings on the planet: animals, plants, bacteria, etc. It is a universal molecule, but one that nevertheless contains genetic information unique to each individual. DNA is thus found in all the cells of living organisms, but each organism also leaves traces of DNA in their environment, through their saliva, gametes, urine, excrement, etc. These fragments of DNA found in nature are called “environmental DNA” or eDNA. They can be detected in water several days after their owner has passed through, while in soil these traces may last for thousands of years.
The environmental forensics
In the same way that forensic scientists have worked out how to decipher the fingerprints we leave behind everywhere we go, fragments of eDNA can tell us a great deal about who left these invisible traces. Each species, in fact, has its own DNA sequence, a little like a genetic “barcode”. The first step in carrying out an eDNA investigation is to collect a sample – a few grams of soil or several litres of water. The DNA is then extracted, amplified and finally sequenced. Bioinformatics tools come into play next, when the DNA sequences are compared with a genetic reference database in order to identify the species present in the study site.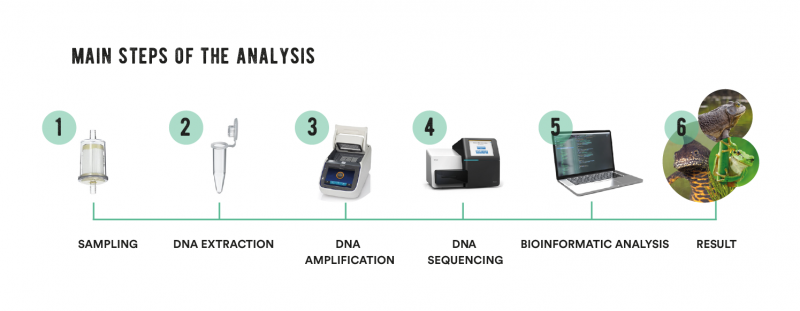 This series of complex tasks requires skill and meticulousness if there is to be any hope of detecting the presence of rare species and avoiding incorrect interpretations. Once mastered, however, this new technology opens up endless possibilities.
This series of complex tasks requires skill and meticulousness if there is to be any hope of detecting the presence of rare species and avoiding incorrect interpretations. Once mastered, however, this new technology opens up endless possibilities.
For example, in the early 2000s, scientists used eDNA in Siberian permafrost to reconstruct ecosystems that dated back tens of thousands of years. The method has also been used to study the diet of endangered species by analysing the genetic material contained in their feces. eDNA is currently revealing its full potential in the building of extensive biodiversity inventories in every aquatic or terrestrial environment.
Towards a paradigm shift?
In the past, to draw up a list of species living in a given area it was often necessary to send numerous specialists covering the various taxonomic groups being studied: fishes, amphibians, mammals, etc. From now on, as long as the genetic “barcode” for each species is known – which requires the considerable expertise of taxonomists – it is possible to conduct a census of the entire biodiversity of an ecosystem from just a single sample.
This is a quick and very effective technique that is often less costly than traditional methods and, most importantly, has no impact on the ecosystem being studied.
A few liters of water from a river, for instance, can be enougth to detect the species of fish living there, along with the amphibians, reptiles, mammals and birds found on its banks. Even the bacteria and viruses that are present but invisible to the naked eye can be detectable. Nevertheless, in order to answer certain ecological questions (population size, gender, stage of development, etc.), the use of traditional inventory methods remains essential.
The highest quality standards
Vigilife’s objective is to standardize the most efficient technologies for monitoring aquatic and terrestrial biodiversity, whether they come from public research laboratories or international private players. The eDNA technologies deployed within the Vigilife framework have been validated by a large number of scientific publications and tested within the framework of numerous scientific projects and expeditions, in order to demonstrate their performance in various ecosystems on a global scale.

journals to validate Vigilife’s eDNA technologies
the most efficient methods in the field
Global biodiversity in Open Data
Thanks to the platform Vigilife Maps, free access to data from eDNA analyses will be quickly made available to environmental managers, researchers, political decision-makers, etc. It will thus be possible to track changes in the distribution and abundance of an endangered species, to be alerted to the appearance of an invasive species, and to study the evolution of biodiversity on a site over time, thanks to synthetic and scientifically validated indicators. These standardized eDNA data will then fill national and international biodiversity databases (GBIF, etc.).
Vigilife is developing a global life monitoring network that will feed national and international databases on biodiversity.
The general public will also be able to learn about the state of health of the ecosystems around them and measure the impact of conservation and restoration programmes carried out by Vigilife partners.

The Vigilife Maps platform, which is the result of a consultation initiated in 2018 between research, conservation and economic actors, will be unveiled soon.
Technology that can be implemented rapidly on a large scale
One of Vigilife’s operational principles is to consolidate the local, national and international databases that exist today thanks to the results of eDNA analyses conducted in the territories. In order to increase the volume and security of data acquisition and analysis from the study sites, expertise resources are gradually being relocated to Vigilife’s partners, as close as possible to the territories. This means every partner being able to process samples in their region and every country being able to maintain sovereignty over its genetic resources. To bring this about, members of the Vigilife alliance are being offered innovative technological solutions, enabling them to carry out eDNA sampling and analysis in a standardised way. These solutions incorporate the highest standards in terms of quality, safety and effectiveness in the field of genetic expertise.
“Plug and play” analysis laboratories
These mobile laboratories have been designed specifically for extracting eDNA. They guarantee the quality of the analyses and also eliminate all risk of contamination of the person carrying out the work or of the environment via pathogens potentially present in the samples being processed.


Optimised sampling methods
Vigilife’s sampling methods have been developed to optimize the detection of all species present on a site, especially the rarest and most difficult to detect. In rivers, for example, they are based on the filtration of large quantities of water (2 X 30 litres), with spatial and temporal eDNA integration.

The development of the first technologies used in the framework of Vigilife (standardized analysis methods, mobile laboratory and mapping platform) was mainly financed by SPYGEN, with the support of the French government (OFB/ADEME – Programme d’Investissements d’Avenir (PIA)/DG Trésor – FASEP), and in collaboration with many international public and private partners.
Our main scientific publications on environmental DNA
-
Deremarque, T. et al. (2024) Ease and Limitations in Using Environmental DNA to Track the Spread of Invasive Host–Parasite Complexes: A Case Study of the Freshwater Fish Pseudorasbora parva and the Cryptic Fungal Parasite Sphaerothecum destruens. Fishes 9, 477
-
Pichot, F. et al. (2024) Mediterranean Islands as Refugia for Elasmobranch and Threatened Fishes. Divers. Distrib. DOI: 10.1111/ddi.13937
-
Feng, K. et al. (2024) Composition, divergence and variability: A comprehensive analysis of fish trait responses to connectivity. Ecol. Indic. 167, 112670
-
Mathon, L. et al. (2024) Three-dimensional conservation planning of fish biodiversity metrics to achieve the deep-sea 30×30 conservation target. Conserv. Biol. DOI: 10.1111/cobi.14368
-
Halabowski, D. et al. (2024) Off the conservation radar: the hidden story of Europe’s tiny pea clams (Bivalvia: Sphaeriidae). Biodivers. Conserv. DOI: 10.1007/s10531-024-02921-x
-
Tourneur, J. et al. (2024) Bivalves des étangs des Espaces naturels sensibles du Maine-et-Loire : diversité et étude pilote sur la détection par analyse de l’ADNe. Naturae 2024
-
Lopes-Lima, M. et al. (2024) Rapid eDNA survey reveals a unique biodiversity hotspot: The Corubal River, West Africa. BioScience DOI: 10.1093/biosci/biae036
-
Jaquier, M. et al. (2024) Environmental DNA recovers fish composition turnover of the coral reefs of West Indian Ocean islands. Ecol. Evol. 14, e11337
-
Haderlé, R. et al. (2024) eDNA-based survey of the marine vertebrate biodiversity off the west coast of Guadeloupe (French West Indies). Biodivers. Data J. 12, e125348
-
Macé, B. et al. (2024) The Tree of Life eDNA metabarcoding reveals a similar taxonomic richness but dissimilar evolutionary lineages between seaports and marine reserves. Mol. Ecol. DOI: 10.1111/mec.17373
-
Manel, S. et al. (2024) Benchmarking fish biodiversity of seaports with eDNA and nearby marine reserves. Conserv. Lett. 17
-
Erős, T. et al. (2024) eDNA metabarcoding reveals the role of habitat specialization and spatial and environmental variability in shaping diversity patterns of fish metacommunities. PLOS ONE 19, e0296310
-
Lefrancois, E. et al. (2024) Validation of an eDNA-based method for surveying fish and crustacean communities in the rivers of the French West Indies. Hydrobiologia DOI: 10.1007/s10750-024-05476-8
-
Baletaud, F. et al. (2023) Comparing Seamounts and Coral Reefs with eDNA and BRUVS Reveals Oases and Refuges on Shallow Seamounts. Biology 12, 1446
-
Veron, P. et al. (2023) Environmental DNA complements scientific trawling in surveys of marine fish biodiversity. ICES J. Mar. Sci. DOI: 10.1093/icesjms/fsad139
-
Cantera, I. et al. (2023) Deforestation strengthens environmental filtering and competitive exclusion in Neotropical streams and rivers. Proc. R. Soc. B 290, 20231130
-
Lamperti, L. et al. (2023) New deep learning‐based methods for visualizing ecosystem properties using environmental DNA metabarcoding data. Mol. Ecol. Resour. DOI: 10.1111/1755-0998.13861
-
Zong, S. et al. (2023) Combining environmental DNA with remote sensing variables to map fish species distributions along a large river. Remote Sens. Ecol. Conserv. DOI: 10.1002/rse2.366
-
Marques, V. et al. (2023) Environmental drivers of eukaryotic plankton and fish biodiversity in an Arctic fjord. Polar Biol. DOI: 10.1007/s00300-023-03187-9
-
Chacko, M.R. et al. (2023) Catchment-based sampling of river eDNA integrates terrestrial and aquatic biodiversity of alpine landscapes. Oecologia DOI: 10.1007/s00442-023-05428-4
-
Rey, A. et al. (2023) Coastal rocky reef fish monitoring in the context of the Marine Strategy Framework Directive: Environmental DNA metabarcoding complements underwater visual census. Ocean Coast. Manag. 241, 106625
-
Mathon, L. et al. (2023) The distribution of coastal fish eDNA sequences in the Anthropocene. Global Ecol Biogeogr DOI: 10.1111/geb.13698
-
Prié, V. et al. (2023) Conservation assessment based on large-scale monitoring of eDNA: Application to freshwater mussels. Biological Conservation 283, 110089
-
Condachou, C. et al. (2023) Inferring functional diversity from environmental DNA metabarcoding. Environ Dna DOI: 10.1002/edn3.391
-
Staentzel, C. et al. (2023) Trophic impact of Neogobius melanostomus in a restored site on the Old Rhine River (France). Aquatic Sciences 85, 46
-
Aucone, E. et al. (2023) Drone-assisted collection of environmental DNA from tree branches for biodiversity monitoring. Sci Robotics 8, eadd5762
-
Coutant, O. et al. (2022) Environmental DNA reveals a mismatch between diversity facets of Amazonian fishes in response to contrasting geographical, environmental and anthropogenic effects. Global Change Biol DOI: 10.1111/gcb.16533
-
Dalongeville, A. et al. (2022) Benchmarking eleven biodiversity indicators based on environmental DNA surveys: More diverse functional traits and evolutionary lineages inside marine reserves. J Appl Ecol DOI: 10.1111/1365-2664.14276
-
Polanco, A. et al. (2022) Ecological indices from environmental DNA to contrast coastal reefs under different anthropogenic pressures. Ecol Evol 12
-
Muff, M. et al. (2022) Environmental DNA highlights fish biodiversity in mesophotic ecosystems. Environ Dna DOI: 10.1002/edn3.358
-
Juhel, J. et al. (2022) Estimating the extended and hidden species diversity from environmental DNA in hyper‐diverse regions. Ecography 2022
-
Pont, D. et al. (2022) Quantitative monitoring of diverse fish communities on a large scale combining eDNA metabarcoding and qPCR. Mol Ecol Resour DOI: 10.1111/1755-0998.13715
-
Hervé, A. et al. (2022) Spatio-temporal variability of eDNA signal and its implication for fish monitoring in lakes. Plos One 17, e0272660
-
Meulenbroek, P. et al. (2022) Sturgeons in large rivers: detecting the near-extinct needles in a haystack via eDNA metabarcoding from water samples. Biodivers Conserv 31, 2817–2832
-
Flück, B. et al. (2022) Applying convolutional neural networks to speed up environmental DNA annotation in a highly diverse ecosystem. Sci Reports 12, 10247
-
Cantera, I. et al. (2022) Low level of anthropization linked to harsh vertebrate biodiversity declines in Amazonia. Nat Commun 13, 3290
-
Sanchez, L. et al. (2022) Ecological indicators based on quantitative eDNA metabarcoding: the case of marine reserves. Ecol Indic 140, 108966
-
Rozanski, R. et al. (2022) Disentangling the components of coastal fish biodiversity in southern Brittany by applying an environmental DNA approach. Environ Dna DOI: 10.1002/edn3.305
-
Mathon, L. et al. (2022) Cross-ocean patterns and processes in fish biodiversity on coral reefs through the lens of eDNA metabarcoding. Proc Royal Soc B 289, 20220162 DOI: 10.1098/rspb.2022.0162
-
Macé, B. et al. (2022) Evaluating bioinformatics pipelines for population‐level inference using environmental DNA. Environ Dna DOI: 10.1002/edn3.269
-
Martin, J. et al. (2021) A comparison of visual observation and DNA metabarcoding to assess the diet of juvenile sea turtle Caretta caretta in the French Mediterranean sea. Mar Freshwater Res DOI: 10.1071/mf21179
-
Bruce, K. et al. (2021). A practical guide to DNA-based methods for biodiversity assessment. , Advanced Books. https://doi.org/10.3897/ab.e68634
-
Cantera, I. et al. (2022) Characterizing the spatial signal of environmental DNA in river systems using a community ecology approach. Mol Ecol Resour DOI: 10.1111/1755-0998.13544
-
Stauffer, S. et al. (2021) How many replicates to accurately estimate fish biodiversity using environmental DNA on coral reefs? Ecol Evol doi:10.1002/ece3.8150
-
Polanco-Fernandez, A. et al. (2021) Detecting aquatic and terrestrial biodiversity in a tropical estuary using environmental DNA. Biotropica DOI: 10.1111/btp.13009
-
Nogueira, J.G. et al. (2021) Alarming decline of freshwater trigger species in western Mediterranean key biodiversity areas. Conserv Biol DOI: 10.1111/cobi.13810
-
Polanco-Fernandez, A. et al. (2021) Comparing the performance of 12S mitochondrial primers for fish environmental DNA across ecosystems. Environ Dna DOI: 10.1002/edn3.232
-
Marques, V. et al. (2021) Use of environmental DNA in assessment of fish functional and phylogenetic diversity. Conserv Biol DOI: 10.1111/cobi.13802
-
Czeglédi, I. et al. (2021) Congruency between two traditional and eDNA-based sampling methods in characterising taxonomic and trait-based structure of fish communities and community-environment relationships in lentic environment. Ecol Indic 129, 107952
-
Leandro, C. et al. (2021) A novel trap design for non-lethal monitoring of dung beetles using eDNA metabarcoding. J Insect Conserv DOI: 10.1007/s10841-021-00329-4
-
Lyet, A. et al. (2021) eDNA sampled from stream networks correlates with camera trap detection rates of terrestrial mammals. Sci Reports 11, 11362
-
Mathon, L. et al. (2021) Benchmarking bioinformatic tools for fast and accurate eDNA metabarcoding species identification. Mol Ecol Resour DOI: 10.1111/1755-0998.13430
-
Boulanger, E. et al. (2021) Environmental DNA metabarcoding reveals and unpacks a biodiversity conservation paradox in Mediterranean marine reserves. Proc Royal Soc B 288, 20210112
-
Coutant, O. et al. (2021) Amazonian mammal monitoring using aquatic environmental DNA. Mol Ecol Resour 21, 1875–1888
-
Prié, V. et al. (2021) Cinq ans d’inventaires des Bivalves de France par analyse de l’ADN environnemental : quelles conclusions, quelles perspectives ? Naturae DOI: 10.5852/naturae2021a8
-
Mena, J.L. et al. (2021) Environmental DNA metabarcoding as a useful tool for evaluating terrestrial mammal diversity in tropical forests. Ecol Appl DOI: 10.1002/eap.2335
-
Marques, V. et al. (2021) GAPeDNA: Assessing and mapping global species gaps in genetic databases for eDNA metabarcoding. Divers Distrib DOI: 10.1111/ddi.13142
-
Juhel, J. et al. (2021) Detection of the elusive Dwarf sperm whale (Kogia sima) using environmental DNA at Malpelo island (Eastern Pacific, Colombia). Ecol Evol DOI: 10.1002/ece3.7057
-
Ficetola, G.F. et al. (2021) Comparison of markers for the monitoring of freshwater benthic biodiversity through DNA metabarcoding. Mol Ecol 30, 3189–3202
-
Milhau, T. et al. (2021) Seasonal dynamics of riverine fish communities using eDNA. J Fish Biol 98, 387–398
-
Pont, D. et al. (2021) The future of fish‐based ecological assessment of European rivers: from traditional EU Water Framework Directive compliant methods to eDNA metabarcoding‐based approaches. J Fish Biol 98, 354–366
-
Juhel, J.-B. et al. (2020) Accumulation curves of environmental DNA sequences predict coastal fish diversity in the coral triangle. Proc Biological Sci 287, 20200248
-
Marques, V. et al. (2020) Blind assessment of vertebrate taxonomic diversity across spatial scales by clustering environmental DNA metabarcoding sequences. Ecography DOI: 10.1111/ecog.05049
-
Polanco-Fernández, A. et al. (2020) Comparing environmental DNA metabarcoding and underwater visual census to monitor tropical reef fishes. Environ Dna DOI: 10.1002/edn3.140
-
Coutant, O. et al. (2020) Detecting fish assemblages with environmental DNA: Does protocol matter? Testing eDNA metabarcoding method robustness. Environ Dna DOI: 10.1002/edn3.158
-
Prié, V. et al. (2020) Environmental DNA metabarcoding for freshwater bivalves biodiversity assessment: methods and results for the Western Palearctic (European sub-region). Hydrobiologia DOI: 10.1007/s10750-020-04260-8
-
Lopes, C.M. et al. (2020) Lost and found: frogs in a biodiversity hotspot rediscovered with environmental DNA. Mol Ecol DOI: 10.1111/mec.15594
-
Dufresnes, C. et al. (2020) Monitoring of the last stronghold of native pool frogs (Pelophylax lessonae) in Western Europe, with implications for their conservation. Eur J Wildlife Res 66, 45
-
Meyer, A. et al. (2020) Morphological vs. DNA metabarcoding approaches for the evaluation of stream ecological status with benthic invertebrates: testing different combinations of markers and strategies of data filtering. Mol Ecol DOI: 10.1111/mec.15723
-
Linard, B. et al. (2020) PEWO: a collection of workflows to benchmark phylogenetic placement. Bioinformatics DOI: 10.1093/bioinformatics/btaa657
-
Spitzen – van der Sluijs, A. et al. (2020) Using environmental DNA for detection of Batrachochytrium salamandrivorans in natural water. Environ Dna DOI: 10.1002/edn3.86
-
Murienne, J. et al. (2019) Aquatic eDNA for monitoring French Guiana biodiversity. Biodivers Data J 7, e37518
-
Vimercati, G. et al. (2019) Assessing the effect of landscape features on pond colonisation by an elusive amphibian invader using environmental DNA. Freshwater Biol 65, 502–513
-
Dufresnes, C. et al. (2019) Early detection and spatial monitoring of an emerging biological invasion by population genetics and environmental DNA metabarcoding. Conservation Sci Pract 1,
-
Miaud, C. et al. (2019) eDNA Increases the Detectability of Ranavirus Infection in an Alpine Amphibian Population. Viruses DOI: 10.3390/v11060526
-
Cantera, I. et al. (2019) Optimizing environmental DNA sampling effort for fish inventories in tropical streams and rivers. Sci Reports 9, 3085
-
Cilleros, K. et al. (2019) Unlocking biodiversity and conservation studies in high‐diversity environments using environmental DNA (eDNA): A test with Guianese freshwater fishes. Mol Ecol Resour 19, 27–46
-
Pont, D. et al. (2018) Environmental DNA reveals quantitative patterns of fish biodiversity in large rivers despite its downstream transportation. Sci Reports 8, 10361
-
Guillerault, N. et al. (2017) Application of DNA metabarcoding on faeces to identify European catfish Silurus glanis diet. J Fish Biol 90, 2214–2219
-
Lopes, C.M. et al. (2017) eDNA metabarcoding: a promising method for anuran surveys in highly diverse tropical forests. Mol Ecol Resour 17, 904–914
-
Sasso, T. et al. (2017) Environmental DNA characterization of amphibian communities in the Brazilian Atlantic forest: Potential application for conservation of a rich and threatened fauna. Biol Conserv 215, 225–232
-
Secondi, J. et al. (2016) Detection of a global aquatic invasive amphibian, Xenopus laevis, using environmental DNA. Amphibia-reptilia 37, 131–136
-
Schneider, J. et al. (2016) Detection of Invasive Mosquito Vectors Using Environmental DNA (eDNA) from Water Samples. Plos One 11, e0162493
-
Leese, F. et al. (2016) DNAqua-Net: Developing new genetic tools for bioassessment and monitoring of aquatic ecosystems in Europe. Res Ideas Outcomes 2, e11321
-
Tesson, S.V.M. et al. (2016) Integrating microorganism and macroorganism dispersal: modes, techniques and challenges with particular focus on co-dispersal. Écoscience 22, 109–124
-
Miaud, C. et al. (2016) Invasive North American bullfrogs transmit lethal fungus Batrachochytrium dendrobatidis infections to native amphibian host species. Biol Invasions 18, 2299–2308
-
Valentini, A. et al. (2016) Next‐generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol Ecol 25, 929–942
-
Civade, R. et al. (2016) Spatial Representativeness of Environmental DNA Metabarcoding Signal for Fish Biodiversity Assessment in a Natural Freshwater System. Plos One 11, e0157366
-
Elbrecht, V. et al. (2016) Testing the potential of a ribosomal 16S marker for DNA metabarcoding of insects. Peerj 4, e1966
-
Bellemain, E. et al. (2016) Trails of river monsters: Detecting critically endangered Mekong giant catfish Pangasianodon gigas using environmental DNA. Global Ecol Conservation 7, 148–156
-
Biggs, J. et al. (2015) Using eDNA to develop a national citizen science-based monitoring programme for the great crested newt (Triturus cristatus). Biol Conserv 183, 19–28
-
Tréguier, A. et al. (2014) Environmental DNA surveillance for invertebrate species: advantages and technical limitations to detect invasive crayfish Procambarus clarkii in freshwater ponds. Journal of Applied Ecology 51, 871–879
-
Giampaoli, S. et al. (2014) The environmental biological signature: NGS profiling for forensic comparison of soils. Forensic Sci Int 240, 41–47
-
Dejean, T. et al. (2012) Improved detection of an alien invasive species through environmental DNA barcoding: the example of the American bullfrog Lithobates catesbeianus. J Appl Ecol 49, 953–959
-
Dejean, T. et al. (2011) Persistence of Environmental DNA in Freshwater Ecosystems. Plos One 6, e23398
-
Bauknecht, R., L. Pellissier, S. Brosse, V. Prié, M. Lopes-Lima, P. Beja, M. K. Goralczyk, A. Polanco Fernandez, J. A. Moreno-Tilano, R. Neme, M. A. Gonzale &, S. Zong 2025.- Combining environmental DNA and remote sensing variables to model fish biodiversity in tropical river ecosystems. Ecological Informatics103251, DOI: 10.1016/j.ecoinf.2025.103251
-
Coutant O, M. Lopes-Lima, J. Murienne, L. Pellissier, G. Quartarollo, A. Valentini, S. Brosse & V. Prié 2025.- No attenuation of fish and mammal biodiversity declines in the Guiana Shield. Science of the Total Environment. 971:179021. DOI: 10.1016/j.scitotenv.2025.179021
-
Nogueira, G. J., A. Lyet, V. Hermoso, P. Beja, M. Lopes-Lima & V. Prié 2025.- Assessing the value of environmental DNA into conservation planning: A case study of freshwater bivalves in France. Journal of Environmental Management 380, 124852. DOI: 10.1016/j.jenvman.2025.124852
-
Lopes-Lima, M., A. Lyet, V. Prié, M. Walters, P. Lindeque, S. M. Kamanja, S. Brosse, F.M.S. Martins, P. Beytell, F. Becker, F. Jacobs, L. Mwapagha, E. C. Fabiano, P. Beja 2025.- A stakeholder empowerment framework to advance eDNA biodiversity monitoring in Africa: Perspectives from Namibia. One Earth8(4) 101244, DOI 10.1016/j.oneear.2025.101244.
-
Prié, V. 2023.- How was France invaded? 170 years of colonisation of metropolitan France by freshwater mussels. Hydrobiologia, DOI: 10.1007/s10750-023-05274-8

































